U-Net: Convolutional Networks for Biomedical Image Segmentation
Abstract
1. Introduction
After the supervised training of a large network with large dataset was adopted to convolutional network, large and deep networks was successful with many visual recognition tasks. Especially in biomedical image processing, the desired output should include localization, and thousands of training images are beyond reach in biomedical tasks. Following Ciresan et al., using a sliding-window setup to predict the class label of each pixel by providing a local region around that pixel, the network can localize, and training data of patches is much larger than the number of training images. However, it is slow since it has to be run separately for each patch, and due to overlapping patches, there is a lot of redundancy. Also, there is a trade-off between localization accuracy and the use of context. Classifier taking into account the features from multiple layers can localize and use the context well at the same time.
Using a fully-convolutional network, that architecture can be extended with requiring few training images and yields more precies segmentations. It replaces pooling operators with upsampling operators, and increase the resolution of the output. Higher resolutioin features from the contracting patch are combined with the upsampled output, therefore increasing the localization. In the upsampling part, feature channels are larger, allowing the network to propagate context informatioin to higher resolution layers. The network uses only the valid part of each convolution, therefore the seamless segmentation of arbitrarily large images by an overlap-tile strategy is available, which uses mirror padding to the border of the image.
To excessively adopt data augmentation, elastic deformatioins to available training images were done, allowing the network learn invatianve to deformations, which is important in biomedical segmentation since deformation used to be the most common variation in real tasks.
Since the separation of touching objects of the same class is the challenging part, use of the weighted loss which heavily weights on separating background labels between touching cells, is proposed in this paper.
2. Network Architecture
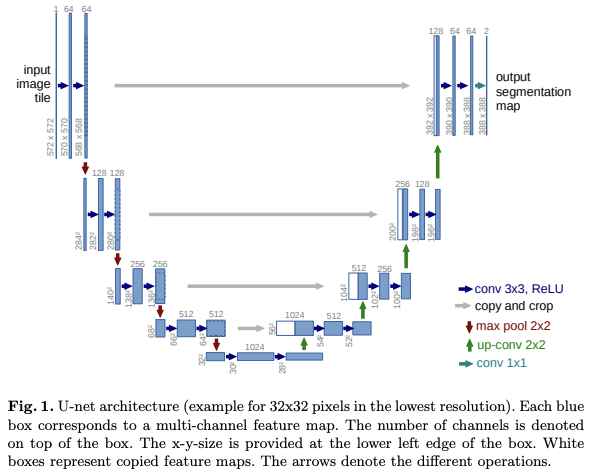
The network is a combination of left contracting part and right expansive part. Repeated two 3x3 convolutions each followed by ReLU and 2x2 maxPool with stride 2 is applied to each layers in downsampling. 2x2 up-convolution followed by two 2x2 convolutions with ReLU is applied for each layer, and final 1x1 convolution for segmentation to map each 64-component feature fecture to desired number of classes is used for last layer in upsampling.
For a seamless tiling of the output segmentation map, it is important to select the right input tile size.
3. Training
The network is trained with SGD of high momentum. The energy function is computed by a pixel-wise softmax over the final feature map combined with the corss entropy loss function. With penalization at each position, it becomes:
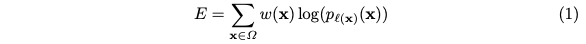
with weight map $w:\Omega\rightarrow\R$ pre-computed for each weight map for each ground truth segmentation, computed as:
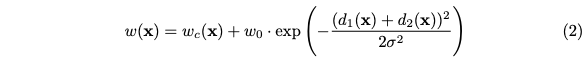
with $d_1:\Omega\rightarrow\R$ the distance to the border of the nearest cell and $d_2:\Omega\rightarrow\R$ the distance to the border of the second nearest cell.
To achieve the initial weights of each feature map in the network approximately unit variance, gaussian distribution with a standard deviation of $\sqrt{2/N}$ is used with N the number of incoming node in the convolution and ReLU layers.
3.1. Data Augmentation
Smooth deformations using random displacement vectors on a coarse 3x3 grid is used. The displacements are sampled from a gaussian distribution, and per-pixel displacements are computed using bicubic interpolation. Dropout is used for end of the each contracting path.
4. Experiments
Experiments done with three tasks: the segmentation of neuronal structures in electron microscopic recordings, cell segmantation task in light microscopic images, and EM segmentation challenge. All of those, the network was above SOTA.
5. Conclusion
WIth the architecture and elastic deformation data augmentation, the performance could exceed that of SOTA.


댓글남기기